One of the surprises (
for me anyway) in discovering the existence of selenocysteine (
Part I. Future Shock and Selenocysteine), was the corresponding discovery that it's encoded by
UGA. Ordinarily, UGA is a stop codon. If a UGA is in an mRNA sequence, it tells the ribosome that the job is done. It's time to pack up all the tRNAs and elongation factors and move on to the next project.
But in the case of selenocysteine, we have a "
work-around." Sometimes UGA stops everything, sometimes the UGA says "put the selenocysteine right here." (
Someone in the office joked that the ID g-o-d must be a programmer since he/she is trying to fix bugs.)
How do the ribosomes and tRNAs know whether to stop or go?They
feel the difference.
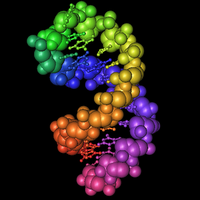
Seriously, the sequences at the 3' end of an mRNA fold into a special hairpin shape like the one shown here (
1,
2,
3). In bacteria, this structure is called a "selenocysteine insertion sequence" or SECIS element. Eucaryotes have similar structures at the 3' ends of mRNAs for selenoproteins.
The RNA in the picture has a rainbow coloring scheme (
Red,
Orange,
Yellow,
Green,
Blue,
Indigo,
Violet). Nucleotides at the 5' end are red, nucleotides at the 3' end are violet. You can follow the colors in the RNA backbone to see how the RNA is twisted around into a hairpin shape.
One of the tests that we used to give job applicants was to have them write a short script for translating a DNA sequence in 6 reading frames. We would give them a mouse pad with the genetic code and put them to work.
Selenocysteine makes this problem a whole lot harder.
Since the recognition feature is a secondary structure, locating the coding sequences for selenocysteines presents an interesting challenge to computational biologists. Finding these sequences requires a bit more than a regular expression.
Kryukov, et. al. describe an algorithm for doing this type of search (
1). They've refined it in the years since this publication, but it seems that the information has yet to percolate through much of the world's bioinformatics community.
And here, I thought I was the only one who seemed to have missed this.Nope.
Can we find selenocysteine in GenBank?I started to wonder if the news about selenocysteine had trickled out beyond PubMed articles and into the rest of the NCBI.
Could I find selenoprotein sequences in the
Gene database? I thought this would be a good place to start since the data are well curated and there are links to reference protein sequences.
I searched and searched, and lo and behold, I found them.

The sequence above codes for human selenoprotein P. U is the one letter symbol that represents selenocysteine. This protein contains an unusually large number of selenocysteines.
I only looked at a few of the reference protein sequences (labeled NP---) from the Gene database, but they all seemed to have selenocysteines.
So the NCBI Gene Database seems to be caught up, at least for the sequences that I checked out.
Mischief and MisannotationsI followed the links to the
Conserved Domain Database. (I'm writing a book on this BTW, and the CDD is really, really cool).
When I got to a summary page, I choose the SelP_C domain (since more U's are on that side of the protein). This gave me a page with a
Pfam alignment between my human SelP sequence and some sequences that were chosen for Pfam. (
you can take a look at this yourself by clicking the link above. Change the format to Hypertext and click Show Alignment to see the selenocysteines in the query sequence).
Reading downward, the Pfam sequences, in the alignment below, are from cow, my query(human), rat, another human sequence, and zebrafish.

Every time my query sequence has a "U," the other sequences have a "c" (
purple boxes above).
This is interesting and odd. Only
one of the proteins with the conserved SelP domain has selenocysteine (and it's our human query sequence).
One interpretation that Kryukov suggested in 2003 (
1), (
and later regretted, I'm sure), is that through evolution, cysteine was substituted for selenocysteine in organisms like the rat and mouse.
I think the presence of the
other human SelP sequence argues for
another interpretation, especially since it's an older version of our query.
If we click the gi links to see the database records, we find something else that's interesting.
A note in the first sequence, from the cow, deposited in April 2006, shows that
someone knew about the selenocysteines,
[MISCELLANEOUS] The selenocysteines are all encoded by the opal codon, UGA.
But apparently, no one bothered to put them in the amino acid sequence, since there aren't any selenocysteines there.
Maybe they didn't read the note.Stranger, yet, the missing selenocysteines could be rationalized away by arguing that the protein sequence is just a conceptual translation - that is, it was determined by using the standard genetic code. Except that using the standard genetic code would have generated
a much shorter sequence since UGA makes translation stop. So, instead of putting in the correct amino acid, the curators (Swiss prot?) typed in the
wrong amino acid. Instead of using the U for selenocysteine, they typed a C for cysteine.
The rat sequence was also updated in April 2006 and we can see that the positions of selenocysteines also seem to be marked in the GenPept record (below), but, funny, there aren't any selenocysteines in the rat sequence.

The other human sequence for SelP and the zebrafish sequence show the same kinds of annotations. Yet
neither one contains selenocysteines in the amino acid sequence.
Could the source of the sequences (Pfam) be the source of the problems?I'm not sure where the problem originates but if I search the
Pfam database at the Sanger Center, for selenocysteine, I get a list of 31 proteins that contain it, and again, I get an annotation that indicates that someone is aware of selenocysteine.
SelP is the only known eukaryotic selenoprotein that contains multiple selenocysteine (Sec) residues...
Yet when I do a seed alignment,
none of the amino acid sequences contain selenocysteine. Here is one example:
SEPP1_HUMAN/22-250 QDQSSLCKQPPAWSIRDQDPMLNSNGSVTVVALLQASCYLCIL
QASKLEDLRVKLKKEGYSNISYIVVNHQGISSRLKYT
The selenocysteines are missing here, too.
If I take this sequence and do a blastp search at the NCBI, I get quite few perfect matches. Just like Pfam, there
are sequences in GenBank that are not yet fixed.
What is our take home message?The simple take-home message, of course, is to be aware the FASTA sequences for selenium-containing proteins
are likely to be wrong. If the annotations say there should be selenocysteine and you can't find a "u" in the sequence, it probably hasn't been fixed yet. Those of us who use the date must always be skeptical and read the literature.
The second take-home message concerns process. The acceptance of new ideas in science generally prompts some re-evaluation of older ideas. We evaluate older concepts more critically in the electric light of new ideas. It would be helpful if these processes could be applied more quickly to sequence data and bioinformatics algorithms. These results support the need for scientific curators who can read the literature, add annotations, and even make corrections in amino acid sequences, from time to time.
The amino acid matrices that we use for protein comparisons, were updated when more sequences became available for doing alignments. We all switched from using PAM to BLOSUM matrices. Maybe it's time to make update the Pfam domains as well.
Selenocysteine exists.
It's time to deal with it and get on with the work.
References:1. Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. 2003.
Characterization of mammalian selenoproteomes.
Science. 300:1439-43.
2. Diamond, AM. 2004.
On the road to selenocysteine. Proc Natl Acad Sci U S A. 101: 13395-13396.
3. Yoshizawa S, Rasubala L, Ose T, Kohda D, Fourmy D, Maenaka K. 2005.
Structural basis for mRNA recognition by elongation factor SelB. Nat Struct Mol Biol. 12:198-203.
Subject: Doing biology with bioinformaticstechnorati tags: bioinformatics, selenocysteine, biochemistry, biology, blast, genetics, genomics, DNA, RNA, Science Education











