Part II. Hunting for huntingtin
In which we're reminded that database searches are experiments, too.
Playing catch-up with the latecomers
Hi, for those of you who've just joined us, we've gotten lost in some databases while hunting for information on huntingtin. If you'd like to catch up a bit and come back later, you might want to read Hunting for huntingtin (part I).
If not, here's a brief synopsis of the plot and what we've done so far:
Then, we got curious about those extra CAGs and wanted to know if they result from the disease or cause the disease. So we looked up huntingtin at the UCSC genome browser and saw that there are similar genes in mouse, pigs, and zebra fish (plus a few other members of the animal kingdom that were not discussed).
Ah hah! Since mice have a similar gene – and we know that the Jackson Lab is the place to go for all things mouse – sure enough, the Jackson mouse breeders have made mice with extra CAGs, and .....the mice get the symptoms of HD.
So you guessed it, the extra CAGS are the problem, not the result.
As the fearless leader of this expedition, I vote now we look at those extra CAGs a little more closely.
Searching for the lost glutamines
You might remember, in part I, I mentioned looking for 3-D structures with polyglutamine. I did find one structure with a polyglutamine sequence, but it looks like the crystallographers weren't able to resolve the part in the structure where the glutamines are supposed to be. Cn3D shows the missing glutamines in grey in the sequence window. The structure window shows this:
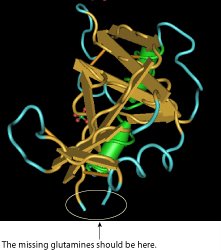
Looking for other structures
Okay, so what can I do? What would you do?
I decided to do a blastp search, since NCBI has this cool new feature where protein sequences, with a corresponding structure, are linked to the structure record in the MMDB.
So I used blastp to search the human protein database with a sequence of 15 glutamines.
What did I find? My result was this: No significant similarity found.
Database searches are experiments, too.
Some of you might be rolling on the floor laughing about now, because you know why I got this kind of result. And some of you might be puzzling over this because it seems kind of contradictory. Didn't I write something earlier about proteins with lots of glutamines? Shouldn't there be some in the database?
Right! Database searches are experiments even if we don't have a pipette anywhere in the room. And there are some fundamental rules for lab experiments that are important to keep in mind.
Okay, Okay, what did we miss?
Close your eyes (oops, no, read the sentence first). Let your mind relax and imagine elementary school science lab. What were the important things to remember about doing experiments?
Disinfect your bench before you start work?
No, not that!
Controls??
Yes.
Experiment Rule 1. Always include control samples, usually both positive and negative.
Personally, I find positive controls to be the most important for database searches. But if I were testing a new program, I would include negative controls, as well.
So I went back and repeated the experiment with a positive control. (I wish this were the only time I've had to do that.) I looked for the huntingtin protein in GenBank and found a reference sequence, NP_002102. And I repeated the blastp search with the reference sequence. (I'm so grateful that I can do this in five minutes on my computer rather than spending a couple of weeks or more trying to get the samples and a couple of days doing a PCR experiment or Southern blot.)
I did a blastp search again. And this time I got a result.
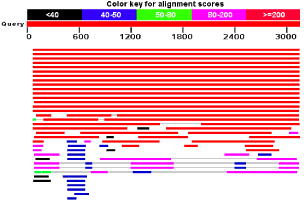
There's definitely something odd about this image.
I'm going to stop writing now and give you a chance to solve the puzzle and find out what's wrong.
And yes, this is one experiment that you can try at home. Take the accession number for huntingtin, NP_002102, type it or paste it into the web form for blastp and see if you can up with answer.
Look for more episodes next week in the continuing saga of the search for huntingtin. In the meantime, feel free to post your guesses or results. I won't give away the answer until next week.
As Tigger says, TTFN!
Read the whole series:
Subject: Doing biology with bioinformatics
Playing catch-up with the latecomers
Hi, for those of you who've just joined us, we've gotten lost in some databases while hunting for information on huntingtin. If you'd like to catch up a bit and come back later, you might want to read Hunting for huntingtin (part I).
If not, here's a brief synopsis of the plot and what we've done so far:
- learned about Woody Guthrie and Nancy Wexler
- found a couple of reviews describing Huntington Disease
- got the HD gene sequence and counted the number of CAGs
- we learned the CAG codes for glutamine and that glutamine can form hydrogen bonds
Then, we got curious about those extra CAGs and wanted to know if they result from the disease or cause the disease. So we looked up huntingtin at the UCSC genome browser and saw that there are similar genes in mouse, pigs, and zebra fish (plus a few other members of the animal kingdom that were not discussed).
Ah hah! Since mice have a similar gene – and we know that the Jackson Lab is the place to go for all things mouse – sure enough, the Jackson mouse breeders have made mice with extra CAGs, and .....the mice get the symptoms of HD.
So you guessed it, the extra CAGS are the problem, not the result.
As the fearless leader of this expedition, I vote now we look at those extra CAGs a little more closely.
Searching for the lost glutamines
You might remember, in part I, I mentioned looking for 3-D structures with polyglutamine. I did find one structure with a polyglutamine sequence, but it looks like the crystallographers weren't able to resolve the part in the structure where the glutamines are supposed to be. Cn3D shows the missing glutamines in grey in the sequence window. The structure window shows this:

Looking for other structures
Okay, so what can I do? What would you do?
I decided to do a blastp search, since NCBI has this cool new feature where protein sequences, with a corresponding structure, are linked to the structure record in the MMDB.
So I used blastp to search the human protein database with a sequence of 15 glutamines.
What did I find? My result was this: No significant similarity found.
Database searches are experiments, too.
Some of you might be rolling on the floor laughing about now, because you know why I got this kind of result. And some of you might be puzzling over this because it seems kind of contradictory. Didn't I write something earlier about proteins with lots of glutamines? Shouldn't there be some in the database?
Right! Database searches are experiments even if we don't have a pipette anywhere in the room. And there are some fundamental rules for lab experiments that are important to keep in mind.
Okay, Okay, what did we miss?
Close your eyes (oops, no, read the sentence first). Let your mind relax and imagine elementary school science lab. What were the important things to remember about doing experiments?
Disinfect your bench before you start work?
No, not that!
Controls??
Yes.
Experiment Rule 1. Always include control samples, usually both positive and negative.
Personally, I find positive controls to be the most important for database searches. But if I were testing a new program, I would include negative controls, as well.
So I went back and repeated the experiment with a positive control. (I wish this were the only time I've had to do that.) I looked for the huntingtin protein in GenBank and found a reference sequence, NP_002102. And I repeated the blastp search with the reference sequence. (I'm so grateful that I can do this in five minutes on my computer rather than spending a couple of weeks or more trying to get the samples and a couple of days doing a PCR experiment or Southern blot.)
I did a blastp search again. And this time I got a result.

or did I??
There's definitely something odd about this image.
I'm going to stop writing now and give you a chance to solve the puzzle and find out what's wrong.
And yes, this is one experiment that you can try at home. Take the accession number for huntingtin, NP_002102, type it or paste it into the web form for blastp and see if you can up with answer.
Look for more episodes next week in the continuing saga of the search for huntingtin. In the meantime, feel free to post your guesses or results. I won't give away the answer until next week.
As Tigger says, TTFN!
Read the whole series:
- Hunting for huntingtin, part I Background, reviews, biochemistry of glutamine, and a bit of comparative genomics
- Hunting for huntingtin, part II In which we're reminded that database searches are experiments, too.
- Hunting for huntingtin, part III Our continuing search for proteins with polyglutamine
- Hunting for huntingtin, part IV: What did you expect to find?
- Hunting for huntingtin, part V: BLASTing on forward
Subject: Doing biology with bioinformatics

0 Comments:
Post a Comment
<< Home