Part. III Hunting for huntingtin
Dealing with distractions
It's been a couple of months since our last installment. "Next week" has come and gone, and like Odysseus on his journey back from Troy, we've experienced delays in getting back to the story. We've likewise faced our own distractions from lotus-eaters, sirens, harpies, the dreaded cyclops. A typical holiday season.
We've even had our own trials with Circe's angry father. If you follow the weather, you know that Neptune unleashed his wrath on the Pacific Northwest this month. Some people tough it out and ride their bikes to work anyway. Most of us just huddle down inside our raincoats, grimly clench our umbrellas and double-tall lattes, and try to hang on until we see sun-breaks or spring.
But I saw the sun today, so it's time to get back to our story.
In our first episode, we learned that Huntington's disease results from the presence of extra CAG's (in the DNA), which are translated to glutamines in the huntingtin protein.
In our second episode, we wanted to know why the extra glutamines were a problem. So we looked for structures with extra glutamines but couldn't find them.
Too many glutamines
Then, we decided to look for other proteins with extra glutamines.
Why?
We looked for two reasons.
First, we could learn something about the structure of polyglutamine (a fancy way of saying lots of glutamines) if we could find a bunch of glutamines in a different protein. Earlier, we wondered if glutamines formed hydrogen bonds with other glutamines or other amino acids. The availability of a different structure might let us test that idea.
Second, we know that people with Huntington's disease get sick because of the extra glutamines. Well, if extra glutamines in the huntingtin protein lead to disease, extra glutamines in other proteins might lead to other genetic diseases. This is just plain interesting stuff to know about.
This is why doing science is kind of like living in "Alice in Wonderland." It's easy to fall down rabbit holes.
Down we go!
Last time, we tried to use blastp to search the protein database for proteins with at least 15 glutamines but couldn't find anything.
So, we searched again with the sequence of the huntingtin protein itself. We know that this entire sequence is in the database and it can serve as a positive control since it has to match itself.
It almost did.
But look at the image. The matching sequences are a bit short on the amino end of the protein sequence (this part of the protein maps towards the 5' end of the mRNA).
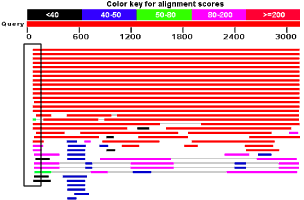 That's odd.
That's odd.
So I looked at the sequence alignments themselves to see what was happening at the amino end of the protein.

The alignment (above) shows that our Query sequence (huntingtin) begins to match a database sequence (identified as Sbjct) at residue 77.
This begs the question: Why isn't there a match to the first 76 amino acids?
So, I looked up the amino sequence for huntingtin in GenBank. Part of the missing section, from 1-71, is shown below. Q stands for glutamine.
matleklmka feslksfqqq qqqqqqqqqq qqqqqqqqpp pppppppppq lpqpppqaqp llpqpqpppp
My conclusion:
The glutamines were missing from the alignment!
Call out John Wayne; it's time for some troubleshooting
We were unable to match glutamines, even with our positive control.
As with any experiment, if the positive control doesn't work, you need to recheck your procedure and find out if something went wrong.
The answer is on the original page of the web form where we began our blastp search.
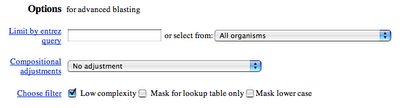 Notice the box that's checked, next to the phrase "Low complexity." The default setting with blastp filters and hides low complexity sequences like pppppp and qqqqqq. In general, this is a good thing, but not when we're trying to find proteins that contain those sequences.
Notice the box that's checked, next to the phrase "Low complexity." The default setting with blastp filters and hides low complexity sequences like pppppp and qqqqqq. In general, this is a good thing, but not when we're trying to find proteins that contain those sequences.
(We use a similar kind of program with DNA sequences, too, called "RepeatMasker.")
Testing the parameters
Let's remove the filter and find out if our positive control (huntingtin) will work, now.
You can check this yourself. Either type or copy and paste the accession number NP_002102 into the blastp web form, and uncheck the low complexity filter before you do the search.
We're on our way. Join us next time, when we do an actual experiment!
Read the whole series:
Subject: Doing biology with bioinformatics
It's been a couple of months since our last installment. "Next week" has come and gone, and like Odysseus on his journey back from Troy, we've experienced delays in getting back to the story. We've likewise faced our own distractions from lotus-eaters, sirens, harpies, the dreaded cyclops. A typical holiday season.
We've even had our own trials with Circe's angry father. If you follow the weather, you know that Neptune unleashed his wrath on the Pacific Northwest this month. Some people tough it out and ride their bikes to work anyway. Most of us just huddle down inside our raincoats, grimly clench our umbrellas and double-tall lattes, and try to hang on until we see sun-breaks or spring.
But I saw the sun today, so it's time to get back to our story.
In our first episode, we learned that Huntington's disease results from the presence of extra CAG's (in the DNA), which are translated to glutamines in the huntingtin protein.
In our second episode, we wanted to know why the extra glutamines were a problem. So we looked for structures with extra glutamines but couldn't find them.
Too many glutamines
Then, we decided to look for other proteins with extra glutamines.
Why?
We looked for two reasons.
First, we could learn something about the structure of polyglutamine (a fancy way of saying lots of glutamines) if we could find a bunch of glutamines in a different protein. Earlier, we wondered if glutamines formed hydrogen bonds with other glutamines or other amino acids. The availability of a different structure might let us test that idea.
Second, we know that people with Huntington's disease get sick because of the extra glutamines. Well, if extra glutamines in the huntingtin protein lead to disease, extra glutamines in other proteins might lead to other genetic diseases. This is just plain interesting stuff to know about.
This is why doing science is kind of like living in "Alice in Wonderland." It's easy to fall down rabbit holes.
Down we go!
Last time, we tried to use blastp to search the protein database for proteins with at least 15 glutamines but couldn't find anything.
So, we searched again with the sequence of the huntingtin protein itself. We know that this entire sequence is in the database and it can serve as a positive control since it has to match itself.
It almost did.
But look at the image. The matching sequences are a bit short on the amino end of the protein sequence (this part of the protein maps towards the 5' end of the mRNA).
 That's odd.
That's odd.So I looked at the sequence alignments themselves to see what was happening at the amino end of the protein.

The alignment (above) shows that our Query sequence (huntingtin) begins to match a database sequence (identified as Sbjct) at residue 77.
This begs the question: Why isn't there a match to the first 76 amino acids?
So, I looked up the amino sequence for huntingtin in GenBank. Part of the missing section, from 1-71, is shown below. Q stands for glutamine.
matleklmka feslksfqqq qqqqqqqqqq qqqqqqqqpp pppppppppq lpqpppqaqp llpqpqpppp
My conclusion:
The glutamines were missing from the alignment!
Call out John Wayne; it's time for some troubleshooting
We were unable to match glutamines, even with our positive control.
As with any experiment, if the positive control doesn't work, you need to recheck your procedure and find out if something went wrong.
The answer is on the original page of the web form where we began our blastp search.
 Notice the box that's checked, next to the phrase "Low complexity." The default setting with blastp filters and hides low complexity sequences like pppppp and qqqqqq. In general, this is a good thing, but not when we're trying to find proteins that contain those sequences.
Notice the box that's checked, next to the phrase "Low complexity." The default setting with blastp filters and hides low complexity sequences like pppppp and qqqqqq. In general, this is a good thing, but not when we're trying to find proteins that contain those sequences.(We use a similar kind of program with DNA sequences, too, called "RepeatMasker.")
Testing the parameters
Let's remove the filter and find out if our positive control (huntingtin) will work, now.
You can check this yourself. Either type or copy and paste the accession number NP_002102 into the blastp web form, and uncheck the low complexity filter before you do the search.
We're on our way. Join us next time, when we do an actual experiment!
Read the whole series:
- Hunting for huntingtin, part I Background, reviews, biochemistry of glutamine, and a bit of comparative genomics
- Hunting for huntingtin, part II In which we're reminded that database searches are experiments, too.
- Hunting for huntingtin, part III Our continuing search for proteins with polyglutamine
- Hunting for huntingtin, part IV: What did you expect to find?
- Hunting for huntingtin, part V: BLASTing on forward
Subject: Doing biology with bioinformatics

0 Comments:
Post a Comment
<< Home